![[IMG]](https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2799835/bin/zpq9990905490001.jpg)
I could not get a larger image to display, as the study is embedded in a technical article. Anyone can follow the link for more details about it. I will post some of the things i found interesting that detailed the findings of the study. I don't have the time to go into detail about it, which would probably bore everyone to tears, anyway. But i'll try to highlight some key points.
The rich fossil record of the family Equidae (Mammalia: Perissodactyla) over the past 55 MY has made it an icon for the patterns and processes of macroevolution. Despite this, many aspects of equid phylogenetic relationships and taxonomy remain unresolved. Recent genetic analyses of extinct equids have revealed unexpected evolutionary patterns and a need for major revisions at the generic, subgeneric, and species levels. To investigate this issue we examine 35 ancient equid specimens from four geographic regions (South America, Europe, Southwest Asia, and South Africa), of which 22 delivered 87–688 bp of reproducible aDNA mitochondrial sequence. Phylogenetic analyses support a major revision of the recent evolutionary history of equids and reveal two new species, a South American hippidion and a descendant of a basal lineage potentially related to Middle Pleistocene equids. Sequences from specimens assigned to the giant extinct Cape zebra, Equus capensis, formed a separate clade within the modern plain zebra species, a phenotypicically plastic group that also included the extinct quagga. In addition, we revise the currently recognized extinction times for two hemione-related equid groups. However, it is apparent that the current dataset cannot solve all of the taxonomic and phylogenetic questions relevant to the evolution of Equus. In light of these findings, we propose a rapid DNA barcoding approach to evaluate the taxonomic status of the many Late Pleistocene fossil Equidae species that have been described from purely morphological analyses.
I am ignoring many of the assumptions of time, macroevolution, & other unsupported assertions in this study, & will focus on the known facts.
- many aspects of equid phylogenetic relationships and taxonomy remain unresolved. That should be obvious. the former definitions, based on 'looks like!' morphologies are often blown up with the hard evidence of genetic lineage. The former lines of equus, popularized in textbooks, nature shows, & slideshows have been pretty much debunked by genetic science.
- a need for major revisions at the generic, subgeneric, and species levels. Clearly. Simply relying on 'looks like' homologies for taxonomic classifications won't do it, anymore. We have hard data, now, with genetic 'markers', & lines that can be followed in the dna.
- the other bolded items say pretty much the same thing. our former beliefs about equus are not accurate. Genetic research has shot some holes in the commonly held beliefs about equus & equidae.
Here is the graphic most of us learned from school & other ToE indoctrination centers:
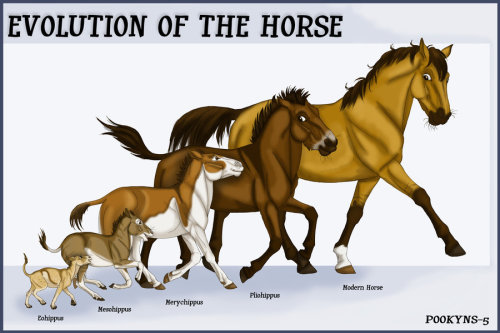
The original linear model of gradual modification of fox-sized animals (Hyracothere horses) to the modern forms has been replaced by a more complex tree, showing periods of explosive diversification and branch extinctions
The 'updated' knowledge about equus is not based on imagined sequences, of purely 'looks like!' descendancy, but has the genetic basis for a family or genus based classification. the first graphic, or those with a circular hub & expanding branches are more accurate, even though the older notions are still promoted as 'settled science!' by many in the ToE indoctrination camp. The earlier belief was a line of evolution, starting with smaller, simpler strains, then getting bigger & more complex. But this is not indicated by the DNA. Many of the formerly held 'ancestors' of equus have been discovered to be not related at all. The imagined sequence of 'evolution!' is only that: Imagined. Here is a more accurate picture of equidae:
![[IMG]](https://www.researchgate.net/profile/Cecilia_Penedo/publication/221755512/figure/fig5/AS:305601142902792@1449872398827/Figure-5-Phylogenetic-tree-of-domestic-horse-breeds-and-Przewalski's-Horse-Parsimony.png)
There is a central, Nuclear genetic type that all other equids come from. They then branch out, diversifying in regions, ecosystems, & climate. But as far as the original ancestor of equidae, not much is known. We can follow the diverse line, but any speculation about the origin of the original equid is just speculation. Here are some key points about equidae:
- All equids are from an original ancestor. They did not originate distinctly from different parent genotypes.
- Equids should ONLY be classified as equids if they can be evidenced to be part of this genetic haplogroup... that is, if they contain the mtDNA marker to indicate descendancy. Big dogs, or other 'looks like a horse!' morphological taxonomies should be discarded in favor of the harder science of genetics.
- Some equids have changed their chromosome numbers, but still can reproduce.. sort of. A donkey with 62 chromosomes can mate with a horse with 64, but produce a sterile mule.
- However, not all odd chromosome matings result in infertile offspring. So there is something else going on to produce a barrier.
- Note in the wiki graphic below that a fox & skunk have the same chromosome count as the horse, but that does not indicate descendancy. The donkey & horse, though, even with different chromosome pairs, have clear evidence of descendancy. IOW, the number of chromosomes is NOT an indicator for evolution or descendancy. It is the MAKEUP of the chromosome that indicates it. The phylogenetic & haplotypes that have the same kinds of genes, structure, & functionality are the indicators, not the number of chromosomes.
- As a reminder, genes, dna, & chromosomes are not like lego blocks, randomly put together in different strands, to make different organisms. Each strand of DNA is unique to the clade it comes from, & can only generate others in the same clade. They can branch out to form different haplotypes, or narrower subsets of the clade, but they are all descended from the same parent stock.
- It is possible that at some time the donkey with its 31 pairs of chromosomes branched off from the horse with its 32 pairs. Chromosomes CAN split & join at the telomere level, but descendancy is still seen in the structure of the 'arm' of the chromosome. Even though there has been some splitting or joining of a chromosome, the basic structure has not changed.. only the length of the telomere, as it has fused or split from the original. All the other genetic information, genes, & structure are the same.. just the connections along the telomere have varied.
Fennec fox
![[IMG]](https://upload.wikimedia.org/wikipedia/commons/thumb/1/1a/Fennec_Foxes.jpg/120px-Fennec_Foxes.jpg) Animals Vulpes zerda 64
Animals Vulpes zerda 64Horse
![[IMG]](https://upload.wikimedia.org/wikipedia/commons/thumb/9/9f/LaMirage_body07.jpg/120px-LaMirage_body07.jpg) Animals Equus ferus caballus 64
Animals Equus ferus caballus 64Spotted skunk
![[IMG]](https://upload.wikimedia.org/wikipedia/commons/thumb/7/7d/Spilogale_gracilis.jpg/120px-Spilogale_gracilis.jpg) Animals Spilogale x 64
Animals Spilogale x 64Mule
![[IMG]](https://upload.wikimedia.org/wikipedia/commons/thumb/e/e0/Juancito.jpg/120px-Juancito.jpg) Animals 63 semi-infertile
Animals 63 semi-infertileDonkey
![[IMG]](https://upload.wikimedia.org/wikipedia/commons/thumb/7/7b/Donkey_1_arp_750px.jpg/120px-Donkey_1_arp_750px.jpg) Animals Equus africanus asinus 62
Animals Equus africanus asinus 62Here is another graphic from the study, showing the descendancy from the mtDNA for the groups sequenced.
![[IMG]](https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2799835/bin/zpq9990905490002.jpg)
They even got a few sequences from extinct genotypes. But they are all descended from the same clade, & their relation is evidenced.
Another interesting point of the study:
"at the molecular level, aDNA studies on a wide range of large mammal taxa (49, 50, 56, 57) have revealed that the loss of genetic diversity over this time period has been much larger than previously recognized"
How is it, that long ago, there was more diversity than now, if the assumption of common ancestry is that new genetic information is constantly being 'created'?
No comments:
Post a Comment